Lab 1
Today we did dissections of several invertebrates.
Jenna and I started with a sea star. Other groups did sea cucumbers, shrimp, urchin, oysters.
Amy and I also dissected a snail. In order to break its shell we had to use the vice in the shop (remember to always go in two's to the shop!)
Lab 2
We worked on prepared histology slides.
Histology is just not my thing!!!
Everything looks like some abstract drawing in pink and purple!
After a lot of work and help from Lisa and Carolyn, I did manage to work my way through some of the abalone's parts. Hopefully I will now always be able to identify the gills, oocytes, mucous and mantle cells. If I am lucky, I will also remember what the hypobranchial gland, the post esophagus and intestine look like... and I will not think that pieces of kelp are part of the tissue :D.
July 25, 2012
Lab 1
Sonia, Jenna, Ashton and I dissected one Armina (sea slug).
Accesion number: 12-1-5
Weight: 43g
Length: 36.4mm
The brain had been previously dissected out by someone else in another lab, and the rest of the organism was donated to us. It had two small lesions. One was a dark lesion on the right side of the lower tip of the foot (5.3mm). It also had a small lesion on left upper side of sea slug. It was kind of hard to see because it was close to where the incision for brain dissection was made.
The digestive gland was extracted and divided into eight parts. Every other slice was set up for histology, and the other 4 pieces where put into a tube with 95% molecular grade ethanol.
Procedure for histology:
- Place in a cassette with the side of interest facing down
- Add a biopsy sponge and close the lid
- Label cassette with the proper accession # using a solvent resistant histology pen
- Place cassette into Invertebrate Davidson’s fixative for 24hr
- After 24 hr, pour off the fixative and replace with 70% ethanol for long-term storage.
July 26, 2012
Lab 1
Today we did DNA extraction. We each worked with our own sample (some used the tissue we dissected out yesterday, and others had samples from 2 years ago).
I worked with the sample we dissected on July 25th (Accesion no. 12-1-5)
We used the Qiagen Stool Kit for DNA Extraction. Almost messed up at the end by adding the wrong buffer, but noticed in time!
Hopefully I will have DNA :D
Lab 2
We prepared the master mixes for the PCR
*Quantities for generic recipes
| Reagent |
uL/reaction |
uL for master mix |
| Immumix 2x |
12.5 |
62.5 |
| BSA |
1.5 |
7.5 |
| Fwd primer |
0.8 |
4 |
| Rev primer |
0.8 |
4 |
| H2O |
7.4 |
37 |
| Template |
2 |
|
| Total |
25 |
I prepared the one for Elrichia primers (EHR16)
*Quantities for special recipe
| Reagent |
ul/reaction |
ul for master mix |
| 5x buffer |
4 |
20 |
| MgCl2 |
1.2 |
6 |
| BSA |
0.8 |
4 |
| H2O |
11.08 |
55.4 |
| dNTP's |
0.4 |
2 |
| Fwd primer |
0.1 |
0.5 |
| Rev primer |
0.1 |
0.5 |
| Taq |
0.32 |
1.6 |
| Template |
2 |
All mixes were distributed. Negative control was added first. We then added Jenna's (forgot to vortex before adding), then mine. Last of all, we added the positive control.
All samples were kept in ice until the rest of the class was finished and the samples could be added to the thermocycler.
* Generic recipe:
| Thermal profile |
Temp |
Time |
| Step1 |
95 |
10min |
| 45 cycles of |
||
| Step 2 |
95 |
15 sec |
| Step 3 |
60 |
1min |
* Special recipe
| Time |
Temp (°C) |
|
| Step 1 |
3 min |
95 |
| Step 2 |
1 min |
95 |
| Step 3 |
30 sec |
62 |
| Step 4 |
30 sec |
72 |
| Repeat steps 2-4, 40 times |
||
| Step 5 |
10 min |
72 |
The thermal profile was left running, and Lisa took out the samples for us and put them in the freezer.
July 27, 2012
Lab 1
We prepared gels for electrophoresis. Everyone in the lab pitched in a bit, and nothing went terribly wrong (some agar was left at the bottom of the flasks, one container was slightly crooked).
We loaded the gels and left them running while we went to lunch.
Lab 2
Here are our results!
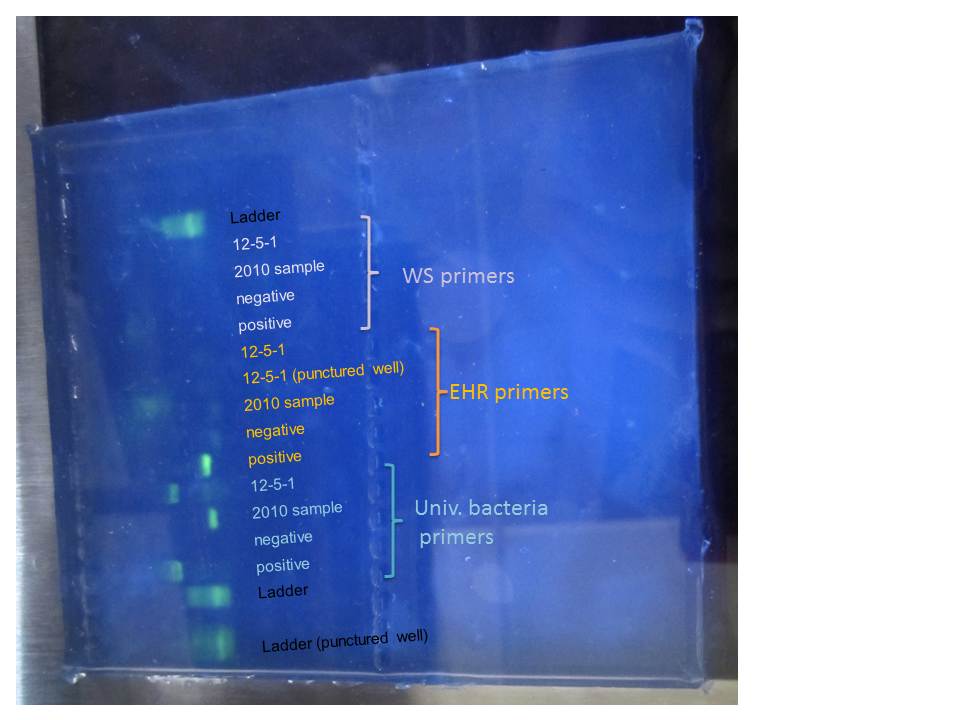 |
| Gel electrophoresis |
The ladders are a bit fuzzy, probably because they were left standing for a little while.
The primers for the universal bacteria worked, though it is funny that the band for the 2010 samples is lower than the positive control and the 12-5-1. We are sure we did not add the wring master mix (but it looks like the band for the positive control in in the EHR mix)
Not much else in the rest of the gel.
Positive control for EHR worked, but not the one for WS. I believe people from the other groups got similar results.
Since this one uses a different recipe from the other two generic ones, there are multiple explanations
- Is some part of the mix not working?
- Was there something wrong in the thermal profile?
July 30, 2012
Lab 1
This morning we went out to False Bay to sample the eel grass. There was a low tide at ~8:15 am so we drove up.
It took a while to set up the transects and organize everyone to start sampling. We started working around 8:50 in the deeper waters, but by that time the tide was already coming back in pretty quickly, so we moved to the shallow end. We got data for 3 transects and went to Drew's house for hot chocolate and tea :D.
Lab 2
We looked at some of the histology slides that were already prepared and tried to identify diseased blades and to count the number of organisms in 5 fields per blade at 40x magnification. However, we could not come to an agreement as to ehat the labys actually look like.
Jenna and I captured all the data that had been recorded in the morning.
We also got some plates running to see if we can culture the labyrinthulids. To di this we
1) Sterilized the area and the dissecting tools
2) Chose the blade to be dissected (I had a healthy one as a control, almost everyone else has diseased ones) and took pictures of it.
3) Cut out the diseased (or in my case, the part that looked the most healthy and had less diatoms) part.
4) Dipped the section for a couple of seconds in 70% ethanol
5) Sterilized tools again and cut the slice of eel grass in 2-3 sections. These were added to culture plates with the media that had been previously prepared by Catherine.
My sample is AEG FB-Sh-Non (transect) Healthy.
In retrospect. I think I might have added too much ethanol to it.
Extra lab
We dissected the diseased urchin than Amy, Jamie and I found when tide-pooling at Lime Kiln Point on Sunday morning.
It was a green urchin, had lost about 1/2 its spines. There was a white edge were the spines were being lost, and the lesions were dark purple with a redish center.
1) Marine agar was prepared by Sammi, Jamie and me, and we innoculated with scrapings from healthy and diseased parts of the test
2) Amy dissected the urchin. She commented that the test was more brittle in the parts were the lesions were greater. As can be seen on the pictures, the inside of the test was a lot darker were the lesions were bigger.
3) We looked at the gonads and the other soft tissue. It looked pretty normal.
4) We also cultured part of the test (dipped into a little bit of ethanol and then cut into smaller pieces).
5) Also cultured some of the liquid that was inside the urchin.
July 31, 2012
This morning we went to the site Beach Comber on Orcas Island to sample the eel grass. We got a lot of data, and it was a lot of fun! I worked with Annie.
Lab 2
We discussed the best way to continue with this case study.
I believe that the best way to proceed would be to first make sure that what we are isolating in culture is in fact Laby, and to really get to now it under the microscope and stuff.
Sonia, Annie and I processed the samples collected today. We also added some healthy blades to dessicant to preserve them for genotypifying later.
Sonia and I replated one of Catherine's plate that had started growing mold. Besides the fungus, there seemed to be two things in there, so we inoculated each in one half of the new culture plate.
Afterwards, I helped Sammi and Jamie look for sea slugs, snails, and eel grass to start an experiment looking at whether these molluscs serve as vectors for laby. I am not confident it will work as it was designed... but who knows, we might get some interesting results.
:D
August 1, 2012
Field 1
Today we went to Picnic Cove to continue with our eel grass study. There was so much to sample! My back was happy when we were finally done. Again, I worked with Annie.
Lab 2
We did DNA extraction for the eel grass samples that Drew and Catherine had collected before.
I performed the extraction on a healthy blade: PC-1H
I helped Courtney and Sammi enter the data that was collected during the day.
August 2, 2012
Field 1
We returned to False Bay to continue sampling there, due to the fact that we were not able to sample completely on Monday because the tide was coming in very quickly. This time we got everything we needed.
Lab 2
We decided to divide forces, so I went to enter data with Courtney, Sammi and Amy. Meanwhile, the rest of the lab prepared everything for PCR. Jenna ran my sample.
August 3, 2012
Lab 1
Courtney, Ashton and I started working on figures from the eelgrass data while the rest of the class prepared everything for the gel electrophoresis. Here are their results:

Comb 1:
1. Ladder
2. Jamie
3. Maya
4. Sonia (2mL) - laby isolate
5. Positive control (2-4, 7)
6. Negative control (2-4, 7)
7. Sonia (5mL) - laby isolate
8. Negative control (9-16)
9. PCH-12
10. PC1-D12
11. PD1
12. PD2
13. LD1
14. LD2
15. LH
16. PH
17. Positive control (9-16)
Comb 2:
1. Ladder
2. Positive control (4-5)
3. Negative control (4-5)
4. Jenna- diseased
5. Ana- healthy
Field
Sammi, Jamie, Jenna, Amy and I went out to get some sand dollars. They were everywhere! Still, it was harder to find healthy ones that diseased ones.
August 4, 2012
Extralab
Jamie, Amy, Sammy, Jenna and I started our side project with the sand dollars
We went through all the sand dollars and selected the ones that looked really healthy (preferably without any signs of predation, though we did need some that had predation but were fully recovered) and the ones that showed the best signs of disease (both bleaching and lesion with algae).
| Top of some really diseased ones that died when transferred to lab |
| Bottom of really diseased ones |
| Three healthy sand dollars |
| Bleached sand dollar. Arrows point to several lesions. |
Transmission experiment:
We set up in boxes with mesh sides and added mud/sand to all of them. Individuals were added as follows (we have pictures of all the individuals):
| Box 3 2 Healthy -51 mm -66 mm 1 Bleached -77 mm |
Box 6- Control 3 Healthy -78 mm -75 mm -58 mm (previous predation) |
Box 2- Healthy baby holding box -22 mm (has lightening to underside) -31 mm -30 mm |
| Box 8 - Control 3 Healthy -48 mm -71 mm (previous predation) -74 mm (previous predation) |
Box 13 2 Healthy - 72 mm -67 mm 1 Bleached -66 mm |
Box 4- Bleached baby box -29 mm -30 mm -26 mm |
| Box 16 2 Healthy -46 mm -68 mm 1 Bleached -89 mm |
Box 14 3 Healthy -72 mm -49 mm -54 mm |
empty |
Culture experiments:
1) We selected the individuals that would be used for the culture and histology, and gave them the following accession numbers:
- SB-1-H: Healthy
- SB-2-B: Bleached-BL
- SB-3-B: Bleached
- SB-4-BL: Bleached and green lesion
- SB-5-BL: Bleached and green lesion, also losing spines and general bad health but still alive.
- SB-6-H: Healthy baby (only used for histology).
| Bleaching |
Lesions |
Histology |
Plates |
Swab |
|
| SB-1-H (large) |
no |
no |
organs |
Marine Agar & TCBS |
Interior and Exterior |
| SB-2-B |
yes |
no |
none |
Marine Agar & TCBS |
Interior and Exterior |
| SB-3-B |
yes |
no |
organs |
Marine Agar & TCBS |
Interior and Exterior |
| SB-4-BL |
yes |
yes |
none |
2x Marine Agar & TCBS lesion and bleaching |
Bleaching Interior and Exterior, Lesion Interior and Exterior |
| SB-5-BL |
yes |
yes |
none |
Marine Agar & TCBS |
Lesion Interior and Exterior |
| SB-6-H (small) |
no |
no |
whole individual |
Marine Agar & TCBS |
Exterior |
Histology:
1) Two bleached, one healthy small sand dollars put in 3.5% formalin, to be transferred to EtOH, then to be decalcified and sectioned and put in cassettes.
2) One healthy (Accession #: SB-1-H), one bleached (SB-3-B) large sand dollars internal organs (gonads, digestive tract) put in cassettes and into Davidson's fixative, to be transferred to EtOH.
->Observations- large bleached sand dollar was male- looked at sperm under microscope. Sex unknown for bleached.
->Gross morphology from dissection- bleached were easier to crack in half, somewhat more brittle test. Healthy specimens interior was clear and dark, diseased interior was cloudy and pale (fluid contained sperm after injuries to organs)
August 6, 2012
Lab 1
We learned how to use the OA lab.
Notes:
-Remember to always rinse electrode, and dry afterwards
-Careful with what you touch with your dirty hands
-Make sure cups are completely dry, and that you dry off whatever is on the walls once you add your sample.
-Small changes in normality of titrator can affect more than the unexact weight of the sample (due to scale)
Lab 2
We looked at all of our sand dollars.
We have vibrio growing on some of the cultures.
I added 1 mL of Laby broth to each well in a 12 well plate so that they can be inoculated tomorrow. It was stored in the fridge.
August 7, 2012
Lab 1
We started working with some microbiology. Each group got a different Vibrio strain.
Jenna and I got RE-101 to work with. We prepared two different plates:
Lab 2
We got baby oysters!
We practiced counting them under the dissecting scope, and estimating the volume that we would have to add to each well in order to have approx. 40 larvae per well.
I think it is easier to count them as they are being suck up by a pipette with a drawn tip (if we do not have to add them to too many wells).
Extralab
We received the histology slides that Ryan sent from SB. They are samples from some of the most common invertebrates there.
We want to be able to tell what eukaryotic parasites are infecting these organisms, and quantify abundance.
August 8, 2012
Lab 1
We did serial dilutions of the Vibrio sample that was given to us.
Jenna and I got RE-98 this time.
This is what we did:
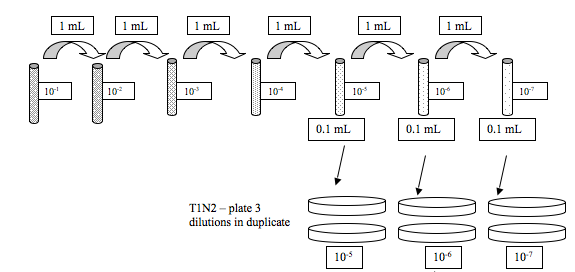
The rest of the group also did serial dilutions for A. media, but we are extra special and did not get plates to do that :)
We also streaked one TCSB plate with our Vibrio sample. All the plates were left for ~2 hrs on the bench before adding parafilm and putting upside down.
Lab2
We prepared the 12-well plates as described in the protocol.About 40 oyster larvae were added to each well. We worked with RE 98 and ambient sea water treatment.
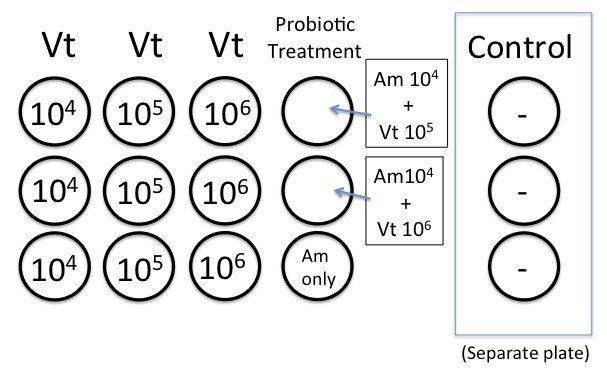
Calculations for the concentrations kind of go as follows:
*Vt after 24 hours: growth at RT (~22C) ~ 3.0 x 109 cfu/mL (from what Elene had seen before)
Because we will add 100 microliters to the well, we will end up with this concentration:
(3.0 x 10^9)(0.1 mL) = 3.0 x 10^8 cfu/mL
3.0 x 10^8 cfu/mL / 4.0 mL SW = 7.5 x 10^7 cfu/mL
original culture:
7.5 x 10^7 cfu/mL
(-1) 1:10 dilution = 7.5 x 10^6 cfu/mL
(-2) 1:10 dilution = 7.5 x 10^5 cfu/mL
(-3) 1:10 dilution = 7.5 x 10^4 cfu/mL
*Am after 24 hr at 37C ~ 1.5 x 10^8 cfu/mL
1.5 x 10^8 x 0.1 = 1.5 x 10^7 cfu/mL
1.5 x 10^7/4.0 = 3.76 x 10^6 / 100 (2 dilutions) = 3.75 x 10^4 cfu/mL
August 9, 2012
Lab 1
No growth was observed on the plates prepared the day before (for the serial dilutions).
The only ones that grew in the group were the ones prepared by Sonia and Ashton.
| 10^-7 |
2 |
2 |
| 10^-6 |
5 |
0 |
| 10^-5 |
32 |
4 |
We counted the oysters that were dead in each of the wells (results are shown below)
Lab 2
We performed Gram stain on Vibrio (gram negative)
I helped Carolyn with alcalinity determinations.
August 10, 2012
Lab 1
We did our larvae count again and bleached them all afterwards to be able to count the total number of larvae per well.
Lab 2
I did alcalinity determinations with Annie while Jenna counted the total oyster larvae in each of our wells.
We also did the azocasein protease assay on our Vibrio strain (positive for protease)
| Well |
Treatment |
9/Aug |
10/Aug |
Total larvae |
| 4 A* |
Control |
0 |
0 |
54 |
| 4 B* |
Control |
1 |
0 |
45 |
| 4 C* |
Control |
0 |
0 |
71 |
| 1 A |
Am |
0 |
6 |
55 |
| 1 B |
Vt 10^6 +Am |
10 |
27 |
54 |
| 1 C |
Vt 10^5 +Am |
14 |
32 |
59 |
| 2 A |
Vt 10^6 |
9 |
29 |
44 |
| 2 B |
Vt 10^6 |
10 |
30 |
54 |
| 2 C |
Vt 10^6 |
13 |
25 |
45 |
| 3 A |
Vt 10^5 |
8 |
30 |
49 |
| 3 B |
Vt 10^5 |
3 |
42 |
58 |
| 3 C |
Vt 10^5 |
5 |
20 |
40 |
| 4 A |
Vt 10^4 |
0 |
18 |
34 |
| 4 B |
Vt 10^4 |
4 |
35 |
42 |
| 4 C |
Vt 10^4 |
1 |
27 |
38 |
August 13, 2012
Using Terminal
Open terminal
Commands
ls
cd: current directory
pwd: present working directory
up arrow instead of retyping everything
You can write cd and then pull the folder you want to work with from the finder.
Cd/Applications/blast/bin
Ls to see what is in that folder and make sure you are where you want
./makeblastdb -in /Users/Shared/EIMD_blast/db/uniprot_sprot.fasta -out /Users/Shared/EIMD_blast/db/uniprot_sprot -dbtype prot
./blastx -query /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta -db /Users/Shared/EIMD_blast/db/uniprot_sprot
Some of us started running a big blast that will be working overnight. this was the command that I put in:
./blastx -query /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta -db /Users/Shared/EIMD_blast/db/uniprot_sprot -outfmt 6 -out /Users/Shared/EIMD_blast/out2/QPX_transcriptome_blastSP -evalue 1E-5 -num_threads 2 -max_target_seqs 1
We started working on My Favorite Gene
August 14, 2012
We finished working on MFG and presented to the class.
We analyzed the QPX. Workflow can be seen below.
August 15, 2012
Here is the summary of what we have done in the past couple of days:
- Trimmed reads [Steven]
- De novo assembly [with CLC-Steven]
- De novo assembly blast against Swiss-Prot [with terminal] (provides just SP ID)
- De novo assembly used to map reads from each library (RNA-Seq) [with CLC-Steven]
- Used Galaxy to get much more information (gene description and GO information) [Galaxy]
- Differentially expressed genes were identified (2-fold) [Excel]
- Used Galaxy to get SP ID of just the Differentially expressed genes [Galaxy]
- __Enrichment Analysis using DAVID__- using Background SP ID and DEG (Gene list) SP ID
- Downloaded files (GO FAT BP) from DAVID
- Tweaked GO FAT BP in Excel
- REVIGO used to visualize Enriched GO BP Processes
Important files for tomorrow:
- Blasting __NCBI blast against nt__ (blast table)
Useful links!!!
Prezi: Great for zooming and animating presentations.
Galaxy: Useful for data transfer, storage, manipulation, bioinformatics.
DAVID: Enrichment analysis
REVIGO: Looking at pretty figures using GO terms from DAVID